| Phone: |
+420 54949 8143, +420 532 234 830 |
| E-mail: |
|
| Office: |
|
Research areas
-
The role of microRNAs/lncRNAs in BCR-signalling and adhesion in B cell malignancies (Chronic Lymphocytic Leukemia, B-Cell Lymphomas)
-
The role of non-coding RNAs in DNA damage response in B cell malignancies (Chronic Lymphocytic Leukemia, B-Cell Lymphomas)
-
Development of novel targeted therapies in B cell malignancies (miR-therapy, targeting of BCR-signalling/adhesion)
-
Prognostic and predictive markers in hematological malignancies
Main objectives
We study the molecular pathways that regulate microenvironmental interactions in normal and malignant immune cells. The microenvironment of immune niches plays an important role in the onset, progression, and resistance of hematological malignancies. We largely focus on the role of non-coding RNAs (microRNAs [miRNAs] and long-noncoding RNAs [lncRNA]) in these pathways. The deregulation of non-coding RNAs can lead to aberrant regulation of pathways that are canonically associated with cancer biology like apoptosis, proliferation and differentiation. However, the role of non-coding RNAs in the microenvironmental interactions of normal and malignant cells is largely unknown.
Content of research
The biology of malignancies derived from „mature“ B cells is largely driven by two molecular pathways that are crucial for the biology of B cells (i) deregulation of B cell receptor signalling, (ii) microenvironment interactions in lymph nodes and bone marrow (reviewed in Seda and Mraz, EJH, 2014)1. These events are interconnected since the normal and malignant B cells are stimulated by antigens and antigen-presenting cells in the context of their microenvironment. Activation of both pathways leads to up-regulation of numerous anti-apoptotic/pro-proliferative molecules and cell adhesion leads to enhanced receptiveness to BCR-signalling and vice versa. It has been shown that these pathways are both important in the onset, progression and resistance of B cell malignancies like chronic lymphocytic leukemia (CLL), follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and mantel cell lymphoma. Targeting microenvironmental interactions in immune niches is currently one of the most promising therapeutic approaches and an important part of the mechanism of action of BCR-inhibitors like ibrutinib or idelalisib (FIG. 1). However, the mechanisms for BCR (de)regulation and mechanism of action of BCR-inhibitors are largely unknown (reviewed in Seda and Mraz, EJH, 2014). Similarly, the role of miRNAs in the regulation of the complex BCR signalling cascade remains one of the highly interesting, yet poorly explored questions, in the biology of normal and malignant B cells.
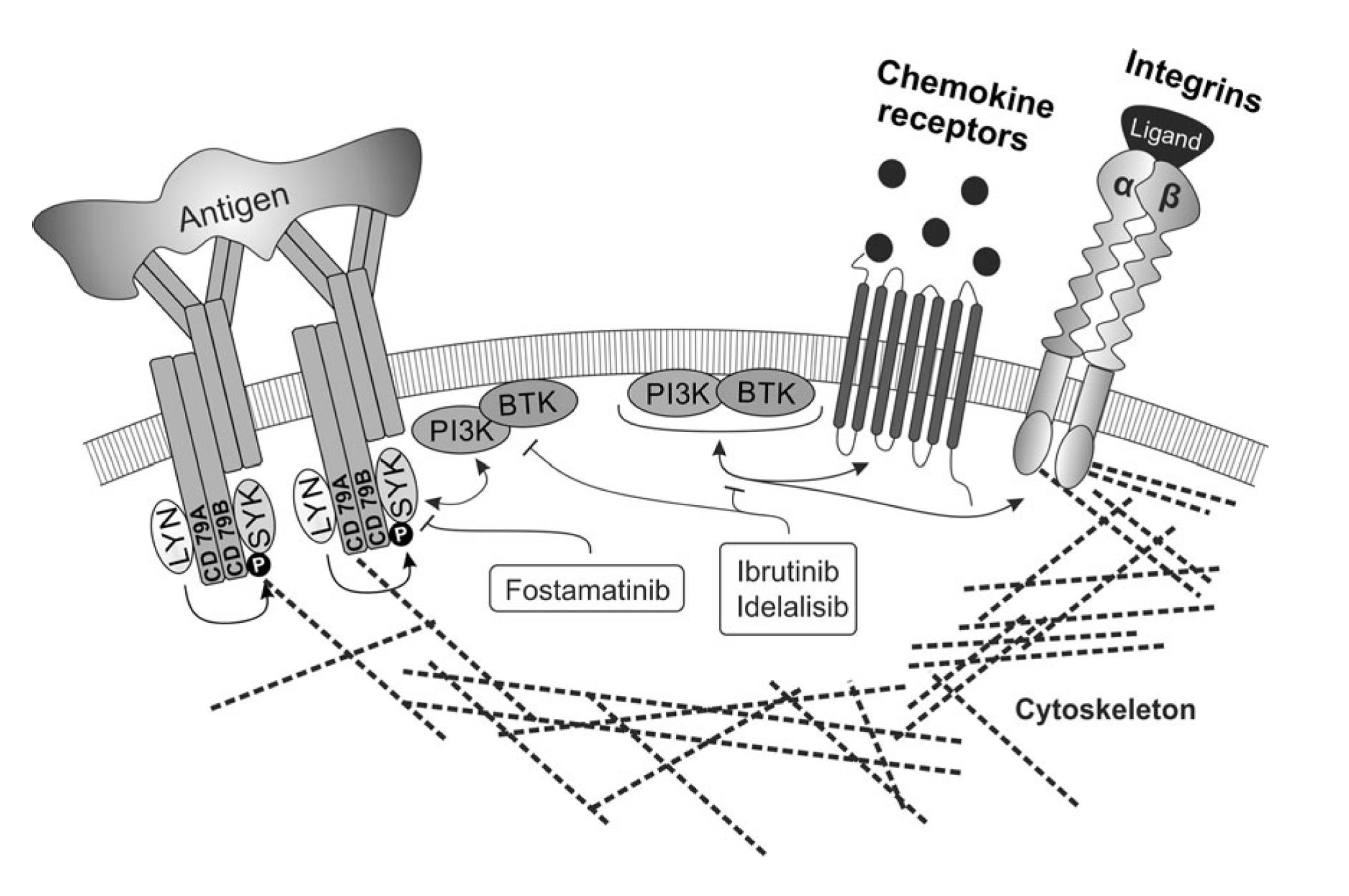
FIG. 1. The crosstalk of BCR signalling and adhesion/migration in B cells, and their therapeutic targeting (Seda and Mraz, EJH, 2014)1
We study molecular pathways involved in microenvironmental interactions in B cell malignancies, and specifically focus on those whose targeting could potentially mimic or synergize with BCR-inhibitors. We mainly focus on “indolent” B cell malignancies (CLL and FL), where there is an unmet clinical need to identify patients with aggressive disease (especially a subset of ~20% of FL patients with unfavorable prognosis) that can benefit from novel and expensive therapeutic approaches, and to describe predictors of response to BCR-inhibitors. We have shown that microRNAs (miRNAs) play a key role in the regulation of microenvironmental interactions, and their expression can be used as prognostic/predictive biomarkers or therapeutically exploited.The discovery of non-coding RNAs, particularly miRNAs, changed the understanding of the biology of normal and malignant B cell and regulation of cell physiology in general. MicroRNAs constitute about 5% of predicted genes in the genome and a single miRNA regulates, based on an incomplete complementarity, many mRNAs which has important consequences for gene regulation. Identifying miRNA targets is not easy since a large number of genes can be influenced by miRNA levels and this is context dependent. MicroRNAs can act as a simple negative regulation of a target mRNA, but it seems that a common scenario is their participation in positive/negative feedback/feed-forward regulatory loops (see Musilova and Mraz, Leukemia, 2015) (FIG. 2). Chronic lymphocytic leukemia (CLL) serves as a prototype malignancy to study the role of miRNAs since it was also the very first human disease associated with deregulation of miRNA expression 8. Several studies including ours have identified ~25 miRNAs differentially expressed between indolent versus aggressive disease (reviewed in Mraz et al., 2009, 2013), including their contribution to p53-pathway deregulation in CLL (Mraz et al., Leukemia, 2009, Mraz et al., Blood, 2012, Mraz et al., Leuk Lymph, 2009). Importantly, we have shown for the first time that miRNAs, namely miR-150, can regulate BCR signalling by directly affecting GAB1 and FOXP1 levels in malignant and normal B cells (Mraz et al., Blood, 2014). We have also shown that miRNAs can target the negative regulators of BCR signalling like SHIP-1 phosphatase (Cui et al., Blood, 2014).
We aim to further describe the role of non-coding RNAs and their targets in the regulation of microenvironmental interactions like adhesion and BCR signalling, and to use this therapeutically.
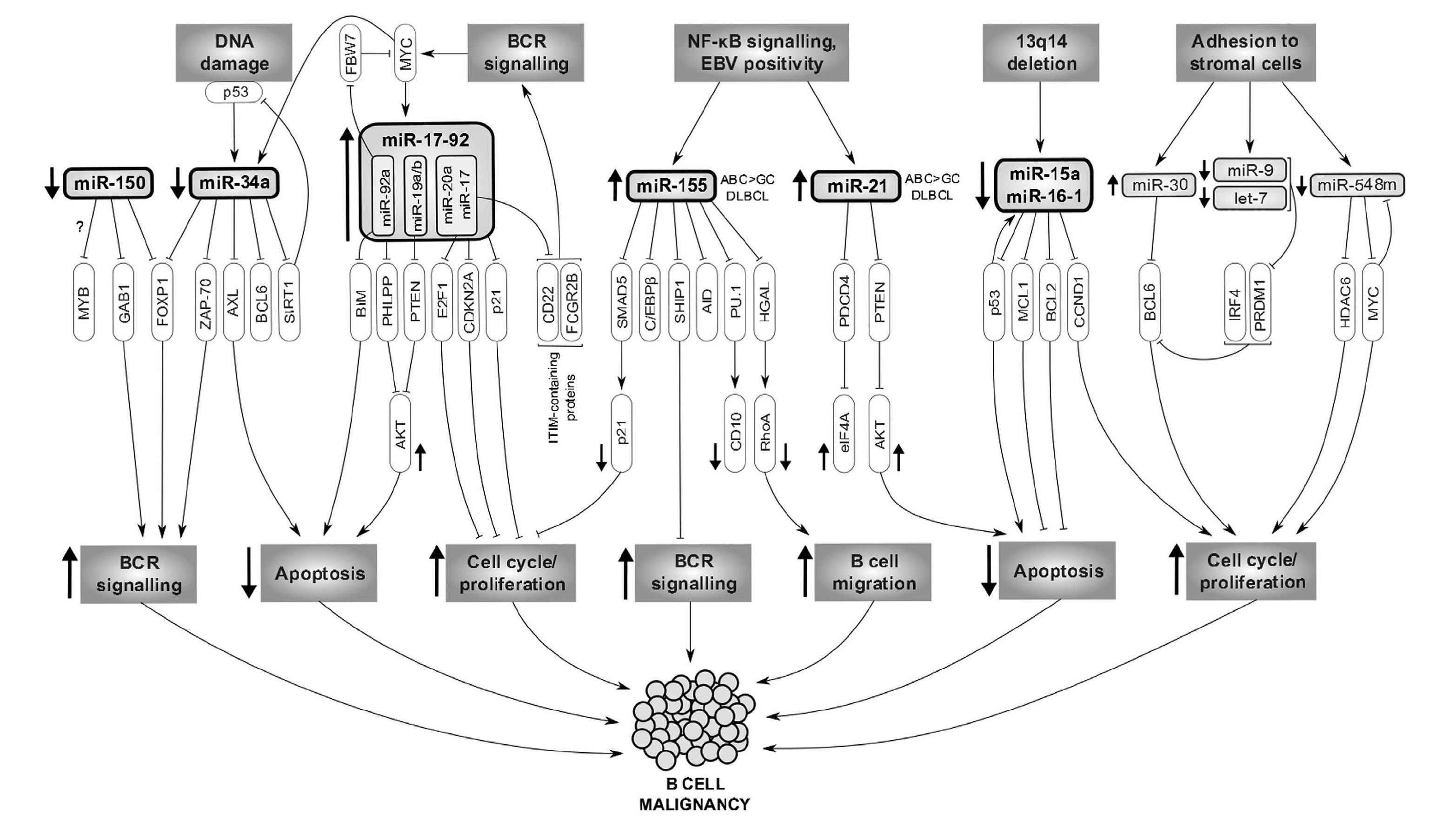
Fig. 2. The contribution of altered miRNA expression to the molecular pathogenesis of B-cell lymphomas. The first row represents the upstream regulation of miRNAs, followed by the primary (third row) and secondary (fourth row) miRNA targets. The last row shows the overall impact of miRNA deregulation on the B-NHL biology (Musilova and Mraz, Leukemia, 2015).
1. Non-Coding RNAS and Microenvironmental Interactions of B-Cell Leukemias and Lymphomas
Supervisor: doc. MUDr. Mgr. Marek Mráz, Ph.D.
Annotation:
The objective of the laboratory is to study the biology of hematological malignancies with a focus on molecular pathways that regulate malignant immune cells’ microenvironmental interactions. It is now understood that pathways like the B-cell receptor (BCR) signaling are deregulated during the onset of most B cell leukemias and lymphomas. The BCR pathway is considered the most promising target for therapy in B cell malignancies. For numerous pathological processes the principal deregulation takes place at the level of such non-coding RNAs in B cell malignancies (Mraz et al., Leukemia, 2009; Mraz et al., Blood, 2012; Musilova and Mraz, Leukemia, 2015). We have recently performed studies which revealed that microRNAs regulate the B-cell receptor signaling which represents the essential pathway for the fate of normal and malignant B cells (Mraz et al., Blood, 2014; Mraz and Kipps, 2013). However, the role of non-coding RNAs in microenvironmental interactions of immune cells is largely unclear. The PhD candidate will perform integrated analysis of coding and non-coding RNAs in the regulation of fundamental microenvironmental interactions of B cell, namely adhesion and BCR signalling. The project will focus on short non-coding RNAs (microRNA) and identification of their targets, but also on long-noncoding RNAs. This will be performed using NGS technics (Illumina) in collaboration with the Genomics Core Facility of CEITEC MU, and experts in NGS data analysis (Uni Torino, CEITEC, EMBL). The candidate will use our in vitro models for microenvironmental interactions, and artificial activation/inhibition of BCR signalling to decipher the regulatory loops that involve coding and non-coding RNAs in primary leukemia cells and in lymphoma cell lines. He will utilize genome editing technics in B cells (using the Crispr/Cas9 technology) to delete miRNAs and test their effect on BCR signalling, activation of kinases and transcription factors. Project will also utilize co-culture of malignant B cells with stromal cells, and/or scaffold materials that mimic human bone marrow, and various other technics of molecular and cell biology (western blotting, cell transfection with miRNA mimics, siRNAs, flow cytometry, cloning etc). Overall, the understanding of microenvironmental interactions is of great interest since it drives the discovery of combinatorial therapy. We will further use the knowledge of gene expression networks to reveal molecular inhibitors that should be used clinically. The interactions within the immune niches are the basis for relapse, persisting minimal residual disease, failure of the immune system to target cancer cells, cell resistance to therapy and, in principle, the incurability of many hematological malignancies.
To apply please contact the supervisor and submit a CV by email to: marek.mraz@email.cz.
Keywords: microenvironmental interactions, B-cell receptor signaling, BCR, non-coding RNAs, microRNA, genome editing, NGS data analysis, leukemia, lymphoma
2. The mechanisms of drug resistance and adaptive response in chronic lymphocytic leukemia
Supervisor: doc. MUDr. Mgr. Marek Mráz, Ph.D.
Annotation
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in the western world with very heterogeneous outcome and unrevealed molecular pathogenesis. It was previously described that patients with the most aggressive disease bear abnormalities of p53 gene or have disrupted DNA damage pathway through other mechanisms (e.g. ATM abnormalities; altered miRNA levels). Despite the progress of novel therapies there are still many resistant cases and patients with very aggressive disease that is not attributed to p53 abnormalities. The identification of mechanisms underlying the resistance and aggressiveness of CLL are of great importance. To study the processes influenced by the most widely used therapy (namely FCR=fludarabine, cyclophosphamide, rituximab), samples collected during the FCR administration will be analyzed on the level of protein-coding and also non-coding genes using the NGS sequencing technology. The aim is to reveal the regulatory circuits responsible for the therapy resistance. The student will also analyze the resistance mechanisms to the novel class of drugs called “BCR signaling inhibitors”. The samples collected during the administration of BCR inhibitors (e.g. Ibrutinib) will be analyzed to understand the mechanisms that leukemic cells utilize to become resistant to therapy. This will be complemented by studies of microenvironmental interactions that influence the adaptation to BCR signaling inhibition or classical chemo-immunotherapy. The project will utilize classical methods of molecular biology (e.g. immunoblotting, flow cytometry, qRT-PCR, cell cultures), and novel Next Generation Sequencing approaches (Illumina) and genome editing (Crispr).
To apply please contact the supervisor and submit a CV by email to: marek.mraz@email.cz.
Keywords: chronic lymphocytic leukemia, CLL, FCR therapy, NGS technology, therapy resistence, BCR signaling inhibitors, genome editing
3. The role of non-coding RNAs in the pathogenesis of aggressive follicular lymphoma and its high-grade transformation
Supervisor: doc. MUDr. Mgr. Marek Mráz, Ph.D.
Annotation
Follicular lymphoma (FL) is a type of blood cancer which originates from B-lymphocytes. It is the most common type of indolent non-Hodgkin lymphoma (NHL) accounting for 20 % of all NHL cases. FL affects mostly older people (median 60 years) and is characterized by slow progression over many years. The prognosis of FL patients is generally good with the median survival around 15 years. However, the clinical course of FL patients can be surprisingly variable (survival from months to decades) and FL still remains incurable. The course of the disease is characterized by repeated relapses leading to the evolution of resistant disease or to the high-grade transformation to a more aggressive diffuse large B-cell lymphoma (DLBCL), which is associated with remarkably poor prognosis and high risk of early death. Number of studies showed that multiple genetic lesions are associated with FL transformation (tFL), however, precise molecular mechanisms underlying this process is largely unclear. Importantly, the role of non-coding RNAs in this process is completely unknown. The aim of the project is to reveal the molecular mechanisms responsible for FL transformation. The primary samples collected before and after high-grade transformation will be analyzed on the level of protein-coding as well as non-coding genes, especially microRNAs (NGS with Illumina). This will be followed by searching for putative targets of tFL-associated miRNAs, and for testing the prognostic significance of relevant miRNAs, long non-coding RNAs and their target mRNAs. This will be also complemented by analysis of genetic aberrations associated with transformed FL. This will help to better understand the disease biology and possibly to identify novel molecular targets that could be used therapeutically. In addition to tFL there is a subset of FL patients who develop early progression or death within 2-3 years following initial diagnosis, and these samples will be analyzed as well. This could identify miRNAs and target mRNAs that can serve as predictors of early relapse after first-line therapy, early transformation, or early death. Classical methodological approaches of molecular biology (immunoblotting, immunostaining, qRT-PCR, cell cultures) will be used together with the novel approaches such as Next Generation Sequencing (Illumina) and genome editing (Crispr).
Keywords: Follicular lymphoma, FL, blood cancer, non-Hodgkin lymphoma, B-cell lymphoma, non-coding RNAs, microRNAs, Next Generation Sequencing, Illumina, genome editing,Crispr
HOW TO APPLY
Candidates should have a master’s degree in Molecular biology, Biochemistry, or similar field and have deep interest in molecular biology and cancer cell biology. Candidate should be willing to collaborate with experts in both wet lab research and bioinformatics. Our institute provides outstanding laboratory facilities and a highly supportive environment. The successful applicant will work in a team of young investigators and will also have the opportunity to supervise bachelor/diploma students. The research will be conducted at CEITEC MASARYK UNIVERSITY (campus Bohunice).
To apply first please contact the supervisor and submit a CV by email to: marek.mraz@email.cz (Subject: PhD School). Information about the laboratory at ceitec.cz/mrazlab.
OTHER INFORMATION
The mission of the institute is to advance the understanding of the underlying molecular and cellular mechanisms of hematological malignancies. Our laboratory is located in the University Campus (Kamenice 5, 4kms outside the city center) and extensively collaborates with the University Hospital Brno in the same campus. The campus provides a vibrant, multidisciplinary and highly collaborative scientific environment. The lab is located in Brno, the second-largest city in Czech Republic that hosts major universities with the biggest concentration of biomedical research in this European region. Brno is one of the major cultural hubs, with a vibrant and lively atmosphere housing ~60000 students. The city has a very good public transport and plenty of interesting places to visit within the reach of trains. Located in the heart of Europe, Brno is within small distance of several major cities (Prague, Vienna, Bratislava, Budapest) and close to international airports (Prague, Vienna).
The NON-CODING AND CODING RNA NETWORKS in the MICROENVIRONMENTAL INTERACTIONS of B-CELL LEUKEMIAS AND LYMPHOMAS
PROJECT:
The objective of the laboratory is to study the biology of hematological malignancies with a focus on molecular pathways that regulate malignant immune cells’ microenvironmental interactions. It is now understood that pathways like the B-cell receptor (BCR) signaling are deregulated during the onset of most B cell leukemias and lymphomas. The BCR pathway is considered the most promising target for therapy in B cell malignancies. For numerous pathological processes the principal deregulation takes place at the level of such non-coding RNAs in B cell malignancies (Mraz et al., Leukemia, 2009; Mraz et al., Blood, 2012; Musilova and Mraz, Leukemia, 2015). We have recently performed studies which revealed that microRNAs regulate the B-cell receptor signaling which represents the essential pathway for the fate of normal and malignant B cells (Mraz et al., Blood, 2014; Mraz and Kipps, Leuk Lymph 2013). However, the role of non-coding RNAs in microenvironmental interactions of immune cells is largely unclear.
The post-doctoral fellow will perform integrated analysis of coding and non-coding RNAs in the regulation of fundamental microenvironmental interactions of B cell, namely adhesion and BCR signalling. The project will focus on short non-coding RNAs (microRNAs) and identification of their targets, but also on long-noncoding RNAs. This will be performed using NGS technics (Illumina) in collaboration with the Genomics Core Facility of CEITEC MU, and experts in NGS data analysis (Uni Torino, CEITEC, EMBL). The candidate will use our in vitro models for microenvironmental interactions, and artificial activation/inhibition of BCR signalling to decipher the regulatory loops that involve coding and non-coding RNAs in primary leukemia cells and in lymphoma cell lines. This will also utilize co-culture of malignant B cells with stromal cells, and/or scaffold materials that mimic human bone marrow. Overall, the understanding of microenvironmental interactions is of great interest since it drives the discovery of combinatorial therapy. We will further use the knowledge of gene expression networks to reveal molecular inhibitors that should be used clinically. The interactions within the immune niches are the basis for relapse, persisting minimal residual disease, failure of the immune system to target cancer cells, cell resistance to therapy and, in principle, the incurability of many hematological malignancies.
HOW TO APPLY:
Candidates should have a PhD or MD with experience in cancer biology or molecular biology (cell culture, cloning technics, WB, flow cytometry), and should possess the ability to conduct independent research. Our institute provides outstanding laboratory facilities (including NGS) and a highly supportive environment. The successful applicant will work in a team of young investigators and will also have the opportunity to supervise doctoral/diploma students. The research will be conducted at CEITEC MASARYK UNIVERSITY (ceitec.eu) in close collaboration with Dept. of Int. Medicine, Hematology and Oncology (University Hospital Brno).
To apply, please submit a CV, a brief statement of research interests, reprints of publications by email to: marek.mraz@email.cz
OTHER INFORMATION:
The mission of the institute is to advance the understanding of the underlying molecular and cellular mechanisms of hematological malignancies. Our laboratory is located University Campus (Kamenice 5, 4kms outside the city center). The campus provides a vibrant, multidisciplinary and highly collaborative scientific environment. The lab is located in Brno, the second-largest city in Czech Rep. that hosts 6 major universities with the biggest concentration of biomedical research in this European region. Brno is one of the major cultural hubs, with a vibrant and lively atmosphere housing ~60000 students. The city has a very good public transport and plenty of interesting places to visit within the reach of trains.
Located in the heart of Europe, Brno is within small distance of several major cities (Prague, Vienna, Bratislava, Budapest) and close to international airports (Prague, Vienna).