| Phone: |
+420 54949 5076 |
| E-mail: |
|
| Office: |
|
Research areas
-
Molecular principles of posttranscriptional regulation of gene expression through alternative splicing, RNA transport and translational control
-
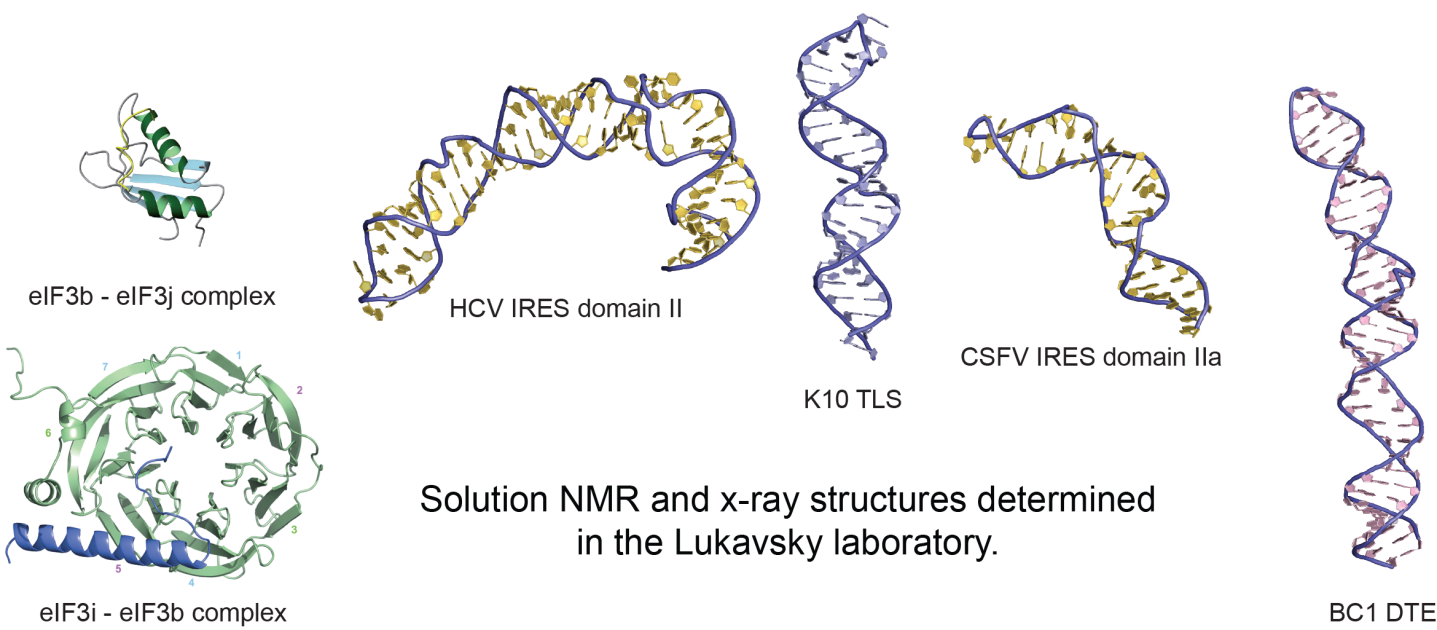
3D structure determination of large RNAs and RNA-protein complexes by solution NMR spectroscopy
-
Biochemical and biophysical studies of RNA-protein interactions
-
Development of novel purification methods and isotope labelling schemes for NMR studies of large RNAs and their assemblies
Main objectives
-
Molecular basis of spatial and temporal control of gene expression through directed RNA transport in dendrites and during development
-
Investigation of molecular principles of RNA-protein interaction networks regulating alternative mRNA splicing of disease-related genes
-
Advancing the size of RNAs and their assemblies amenable to solution NMR studies
Content of research
Post-transcriptional regulation of gene expression is based on regulatory RNA elements which determine the protein sequence of a gene product through pre-mRNA alternative splicing in the nucleus and control temporal and spatial pattern of protein synthesis in the cytoplasm. While we begin to decipher the splicing code, only very little is known about how mRNAs reach their cellular compartments to ensure that proteins are expressed in the right place at the right time within the cell. This compositional, spatial and temporal control of gene expression is key for maintaining the order of events during development and for local response to stimuli in specialized cells such as neurons and, thus, deregulation often leads to human disease.
Our aim is to unravel molecular principles governing post-transcriptional regulation of gene expression. Using NMR spectroscopy as our main structural tool, we will study RNA-protein (RNP) interaction networks regulating alternative splicing of disease-related genes and RNA elements and their protein assemblies crucial for control of protein synthesis in the cytoplasm.
At CEITEC, we are in a good position to tackle these large biological RNP systems with the excellent high-field NMR equipment in place and since we pioneered NMR structure determination of large RNA systems in the past. Our NMR structural work is always complemented with biochemistry, x-ray crystallography, other biophysical methods and cell biology to create a comprehensive molecular description of RNP function. Failure to properly splice mRNAs and to transport them into the right cellular compartment can lead to impairment of memory formation and severe human diseases. Molecular insights into mechanisms governing RNA-based regulation of gene expression is therefore fundamentally important for understanding neurobiology, neurological disorders and human disease, and is key for the development of successful therapies in the future.
CURRENT RESEARCH INFRASTRUCTURE
The current NMR laboratory at CEITEC hosts six high-field NMR instruments ranging from 500 to 950 MHz equipped with cryoprobes for spectroscopy in liquids. The lab also provides state-of-the-art instrumentation for biochemical and biophysical characterization of RNA-protein interactions, protein and RNA preparation and purification, and computational tools for biomolecular structure determination.
1. The Three Prime Untranslated Region (3'UTR)
Supervisor: Mgr. PharmDr. Peter Lukavsky
Consultants: MVDr. Boris Tichý, Ph.D., doc. RNDr. Zbyněk Zdráhal, Dr.
Annotation:
The fate of each mRNA and thus its protein product is highly dependent on its 3’ untranslated region (3’UTR) which encodes spatial and temporal signals for gene expression. In higher eukaryotes and especially in neurons, long 3’UTRs (up to 14 kb) contain numerous conserved sequence elements but most of their functions are unknown. These conserved RNA elements associate with multiple protein partners to form large RNA-protein assemblies, so-called mRNPs which affect mRNA stability, mediate directed mRNA transport along the cytoskeleton or regulate translation efficiency. Thus the mRNPs determine when and where and how much of a protein product is made. As a consequence, it is not surprising that 3’UTR mutations and length alterations change the spatial and temporal pattern of gene expression which can create imbalances and thus diseases, such as cancer, heart disease and neurodegeneration. In this project, we want to deepen our molecular understanding of the regulatory 3’UTR RNA-protein networks. We propose to study secondary structure, composition and tertiary structure of functional units of the mRNP assembled on the CaMKIIa 3’UTR, a dendritic mRNA implicated in learning and memory formation. The goal is to characterize protein-RNA interactions in the 3 kb long 3’UTR and to assign the cellular functions of these units using state-of-the-art techniques, such as SHAPE sequencing, bioinformatics, RNA-based pulldown, proteomic analysis, functional in vivo assays and structural studies. Our results aim to provide the first comprehensive description of 3’UTR structure and function. This will lead to a better understanding of disease-related 3’UTR mutations and length alterations and their effect the spatio-temporal control of gene expression.
Keywords: Three Prime Untranslated Region, 3’UTR, mRNPs, RNA-protein networks, CaMKIIa, SHAPE sequencing, RNA-based pulldown, gene expression, 3’UTR mutations