| Phone: |
+420 54949 2436 |
| E-mail: |
|
| Office: |
|

| Phone: |
+420 54949 3388, +420 54949 5507, +420 54949 3253 |
| E-mail: |
,
|
| Office: |
|
Research areas
-
Protein-RNA and protein-protein interactions and their roles in the regulation of gene expression
-
3’-end processing and transcription termination
-
NMR spectroscopy of proteins and nucleic acids and their complexes
-
Development and application of new methods to aid the interpretation of NMR data
Main objectives
-
Investigation of the role of RNA in gene expression, development and human diseases.
-
Establishing an isotope laboratory for NMR studies and the development of new strategies for the preparation of isotopically labelled proteins in eukaryotic cells.
Content of research
RNA is essential for cell survival. Not only is it a messenger between the genomes and proteomes but it also carries out, or participates in, many functions such as RNA processing and protein translation, acting as structural scaffolds, transporters, gene regulators and biocatalysts. We will help to clarify molecular mechanisms underlying RNA quality control in eukaryotic cells through the investigation of the detailed biochemical principles of RNA recognition, processing and degradation. We will use a combination of biochemical, genetic, and structural methods to unravel the molecular mechanism of eukaryotic RNA surveillance.
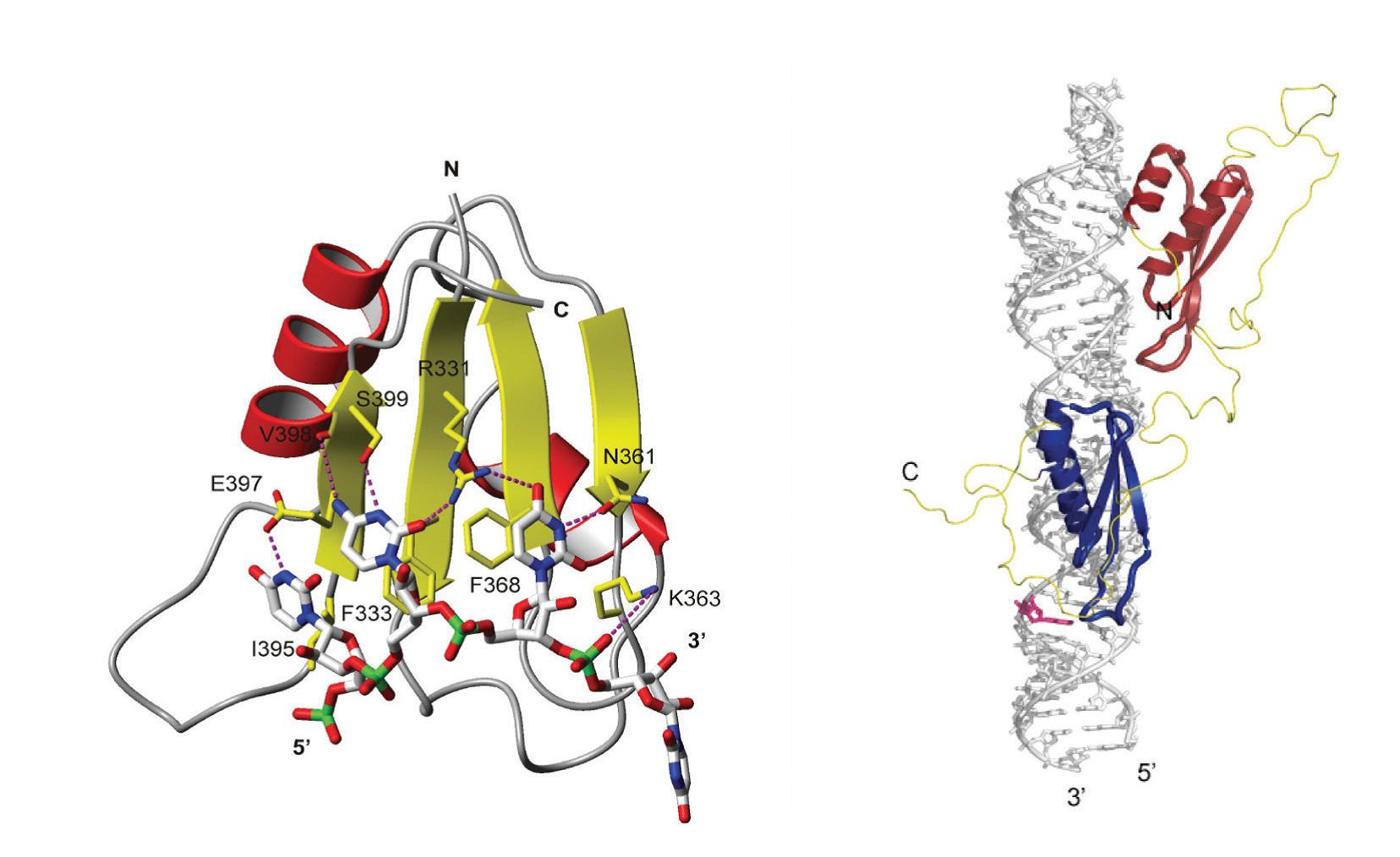
Example of NMR spectroscopy investigation of RNA sequence- and structure-dependent recognition by proteins.
1. A structural basis for the cross-talk between histones and RNA Polymerase II
Supervisor: doc. Mgr. Richard Štefl, Ph.D.
Consultants: Mgr. Karel Kubíček, PhD., doc. Mgr. Štěpánka Vaňáčová, Ph.D.
Annotation:
Chromatin is a highly flexible architecture in which spatially and temporally coordinated changes between structurally condensed states (transcriptionally repressive), and structurally accessible states (transcriptionally active) regulate gene expression. Posttranslational modifications of histones play the fundamental role in maintaining the dynamic equilibrium of these two chromatin states. Individual histone modifications are associated with a given stage of chromatin remodelling and transcriptional cycle. Interestingly, the C-terminal domain (CTD) of RNA polymerase II (RNAPII) is also posttranslationally modified which serves as a signal for the recruitment of appropriate processing factors in coordination with the transcription cycle. Recent findings revealed that these is an overlap between the occurrence of the histone and CTD marks, suggesting the existence of a cross-talk between the chromatin remodelers (orchestrated by histone modifications) and transcription/processing factors (orchestrated by RNAPII CTD modifications). We will identify protein adaptors that spatially and temporally mediate interactions between chromatin and RNAPII. These proteins will be structurally characterized and we will reveal the structural basis for the cross-talk between histones and RNAPII that is orchestrated through their posttranslational modifications.
Keywords: RNA, posttranslational modifications, CTD, RNAPII, histones, chromatin, C-terminal domain, gene expression
2. Cracking the CTD code
Supervisor: doc. Mgr. Richard Štefl, Ph.D.
Consultants: Mgr. Karel Kubíček, PhD., Mgr. Pavel Plevka, Ph.D.
Annotation:
The concept of the CTD code that specifies the position of RNAPII in the transcriptional cycle and thus recruits specific processing factors, was suggested almost a decade ago. However, how the C-terminal domain (CTD) of RNA polymerase II recruits, activates, and displaces appropriate processing factors in coordination with the transcription cycle, remains obscure. Association of specific factors with the CTD is dictated by different post-translational modification patterns and conformational changes in the CTD. To reveal the CTD code, a combination of structural, biochemical, and genetic methods will be used to study important complexes of the CTD with CTD code reader proteins. The project will help to decipher the basic rules that govern the readout of the CTD code.
Keywords: CTD, TNAPII, C-terminal domain, CTD code, post-translational modification